library(tidyverse)
theme_set(theme_classic() +
theme(panel.grid.major.y = element_line(color = "grey92")))
Bayes’s Theorem
The Bayes’s theorem is, surprisingly (or unsurprisingly), very simple:
P(B∣A)=P(A∣B)P(B)P(A)
More generally, we can expand it to incorporate the law of total
probability to make it more applicable to data analysis. Consider Bi as one of the n many possible mutually exclusive
events, then P(Bi∣A)=P(A∣Bi)P(Bi)P(A)=P(A∣Bi)P(Bi)P(A∣B1)P(B1)+P(A∣B2)P(B2)+⋯+P(A∣Bn)P(Bn)=P(A∣Bi)P(Bi)∑nk=1P(A∣Bk)P(Bk)
If Bi is a continuous variable,
we will replace the sum by an integral, P(Bi∣A)=P(A∣Bi)P(Bi)∫kP(A∣Bk)P(Bk)
P(Bi∣A)∝P(A∣Bi)P(Bi)
Example 1: Base rate fallacy (From Wikipedia)
A police officer stops a driver at random and does a breathalyzer test for the driver. The breathalyzer is known to detect true drunkenness 100% of the time, but in 1% of the cases, it gives a false positive when the driver is sober. We also know that in general, for every 1,000 drivers passing through that spot, one is driving drunk. Suppose that the breathalyzer shows positive for the driver. What is the probability that the driver is truly drunk?
P(positive|drunk)=1
P(positive|sober)=0.01
P(drunk)=1/1000
P(sober)=999/1000
Using Bayes’ Theorem,
P(drunk|positive)=P(positive|drunk)P(drunk)P(positive|drunk)P(drunk)+P(positive|sober)P(sober)=1×0.0011×0.001+0.01×0.999=100/1099≈0.091
So there is less than a 10% chance that the driver is drunk even when the breathalyzer shows positive.
You can verify that with a simulation using R:
#> truly_drunk
#> drunk sober
#> 100 99900breathalyzer_test <- ifelse(truly_drunk == "drunk",
# If drunk, 100% chance of showing positive
"positive",
# If not drunk, 1% chance of showing positive
sample(c("positive", "negative"), 999000,
replace = TRUE, prob = c(.01, .99)
)
)
# Check the probability p(positive | sober)
table(breathalyzer_test[truly_drunk == "sober"])
#>
#> negative positive
#> 98903 997# 997 / 99900 = 0.00997998, so the error rate is less than 1%
# Now, Check the probability p(drunk | positive)
table(truly_drunk[breathalyzer_test == "positive"])
#>
#> drunk sober
#> 100 997# 100 / (100 + 997) = 0.0911577, which is only 9.1%!
Bayesian Statistics
Bayesian statistics is a way to estimate some parameter
θ (i.e., some quantities of
interest, such as the population mean, regression coefficient, etc) by
applying the Bayes’ Theorem. > P(θ|D)∝P(D|θ)P(θ)
There are three components in the above equality:
- P(D|θ), the probability
that you observe data D, given the
parameter θ; this is called
the
likelihood(Note: It is the likelihood of θ, but probability about y) - P(θ), the probability
distribution θ, without
referring to the data D. This
usually requires appeals to one’s degree of belief, and so is called the
prior - P(θ|y), the updated
probability distribution of θ,
after observing the data D; this is
called the
posterior
On the other hand, classical/frequentist statistics focuses solely on
the likelihood function.
Example 2: Locating a Plane
Consider a highly simplified scenario of locating a missing plane in the sea. Assume that we know the plane, before missing, happened to be flying on the same latitude, heading west across the Pacific, so we only need to find the longitude of it. We want to go out to collect debris (data) so that we can narrow the location (θ) of the plane down.
We start with our prior. Assume that we have some rough idea that the plane should be, so we express our belief in a probability distribution like the following:
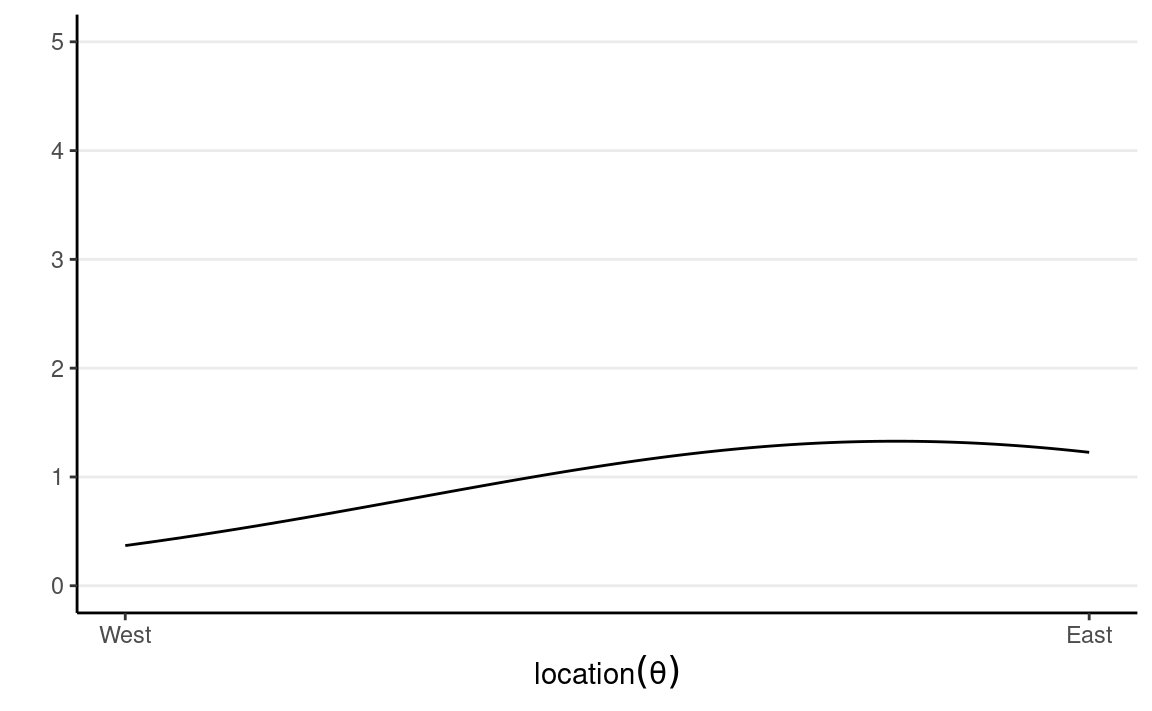
which says that our belief is that the plane is about twice more likely to be towards the east than towards the west. Below are two other options for priors (out of infinitely many), one providing virtually no information and the other encoding stronger information:
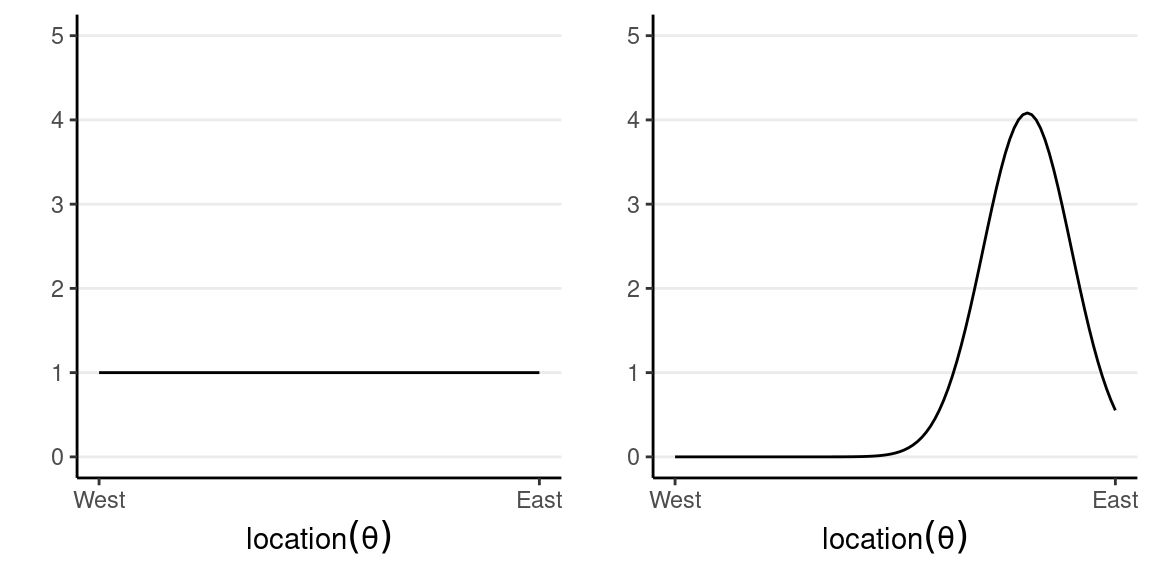
The prior is chosen to reflect the researcher’s belief, so it is likely that different researchers will formulate a different prior for the same problem, and that’s okay as long as the prior is reasonable and justified. Later we will learn that in regular Bayesian analyses, with moderate sample size, different priors generally make only negligible differences.
Now, assume that we have collected debris in the locations shown in the graph,
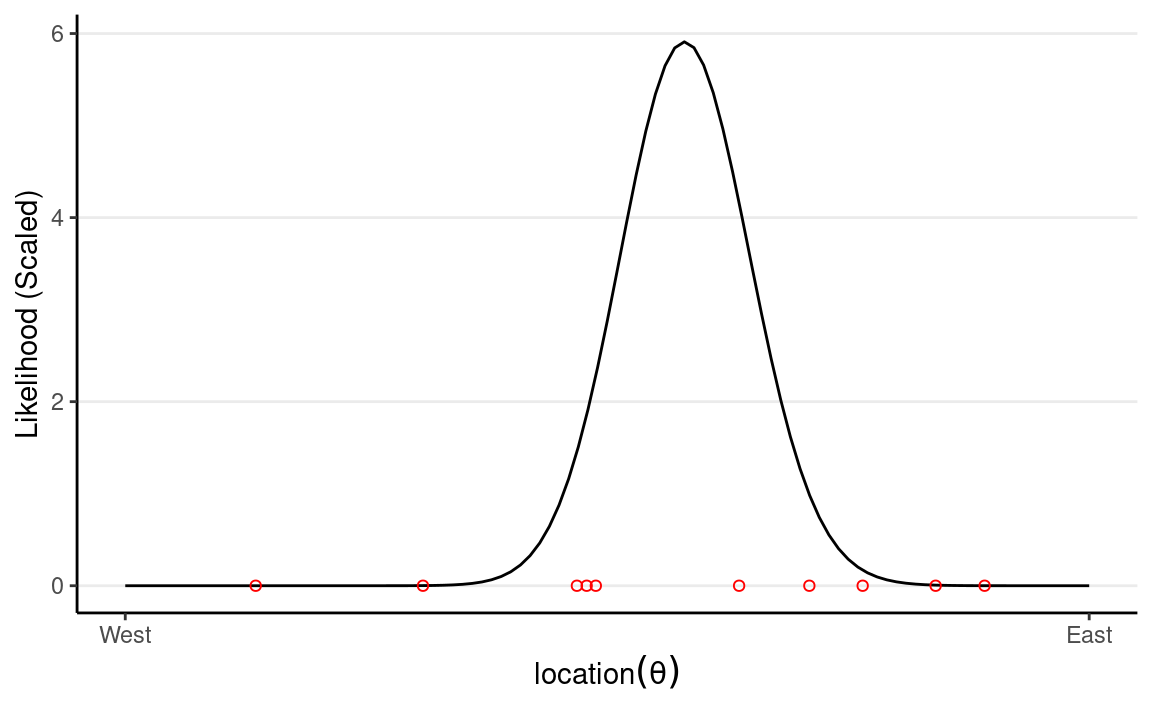
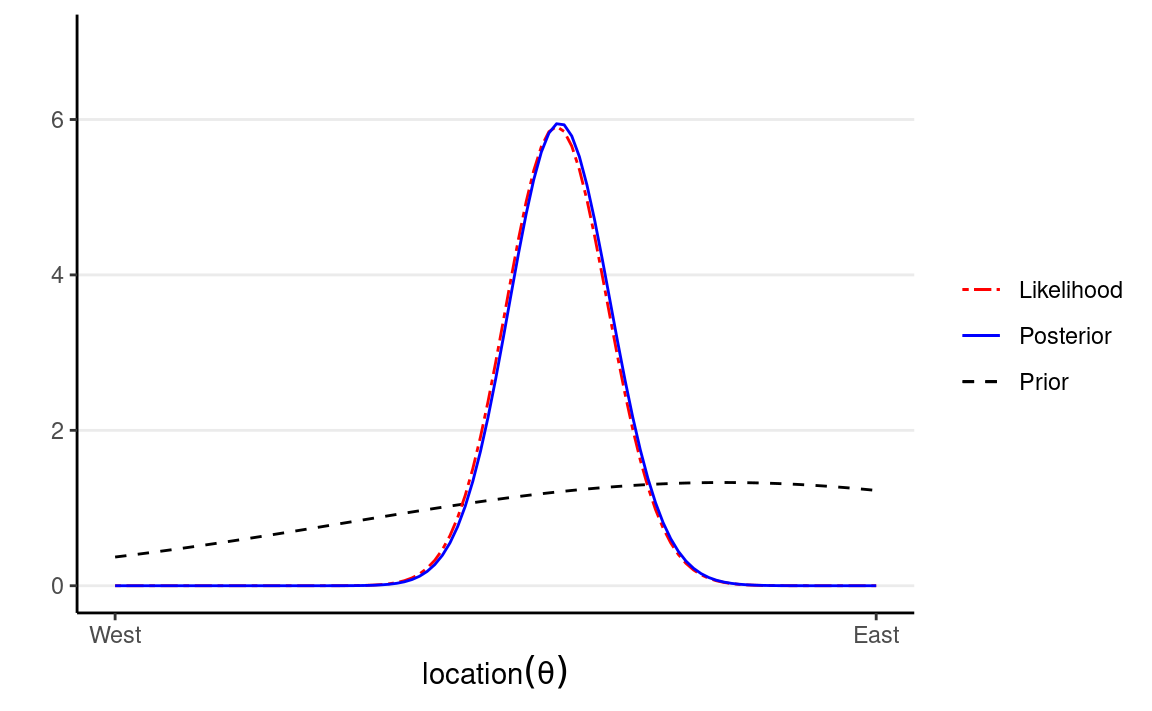
Now, from Bayes’s Theorem,
Posterior Probability∝Prior Probability×Likelihood
So we can simply multiply the prior probabilities and the likelihood to get the posterior probability for every location. A rescaling step is needed to ensure that the area under the curve will be 1, which is usually performed by the software.
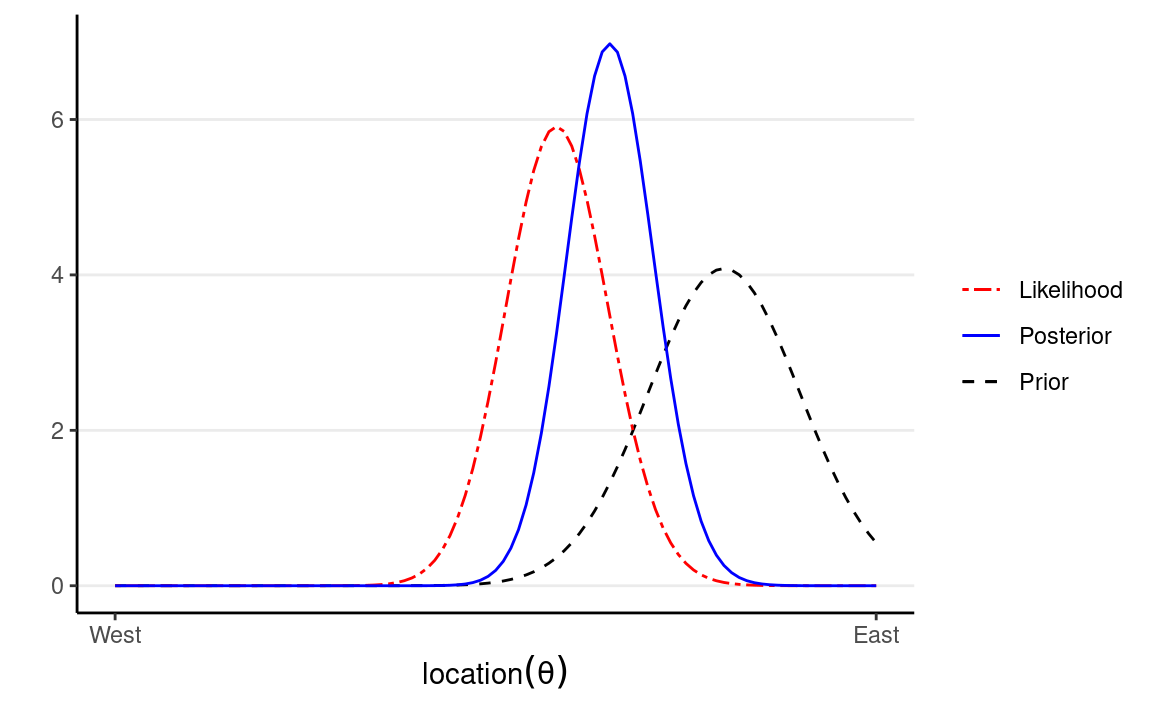
Influence of Prior
The following shows what happen with a stronger prior:
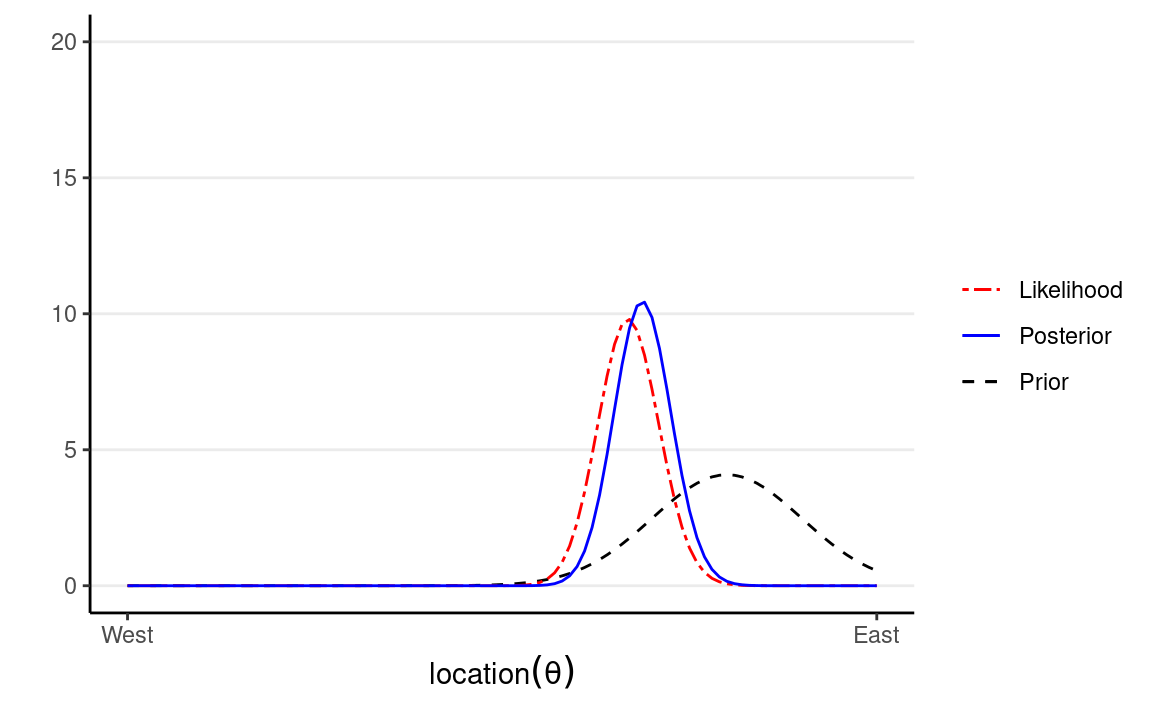
Influence of More Data
The following shows what happen with 20 more data points:
Data-Order Invariance
In many data analysis applications, researchers collect some data D1, and then collect some more data D2. An example would be researchers conducting two separate experiments to study the same research question. In Bayesian statistics, one can consider three ways to obtain the posterior:
- Update the belief with D1, and then with D2
- Update the belief with D2, and then with D1
- Update the belief with both D1 and D2 simultaneously
Whether these three ways give the same posterior depends on whether
data-order invariance holds. If the inference of D1 does not depend on D2, or vice versa, then all three ways
lead to the same posterior. Specifically, if we have conditional
independence such that P(D1,D2∣θ)=P(D1∣θ)P(D2∣θ),
Exchangeability*
Exchangeability is an important concept in Bayesian statistics. Data are exchangeable when the joint distribution, P(D1,…,DN), does not depend on the ordering of the data. A simple way to think about it is if you scramble the order of your outcome variable in your data set and still can obtain the same statistical results, then the data are exchangeable. An example situation where data are not exchangeable is
- D1 is from year 1990, D2 is from year 2020, and the parameter θ changes from 1990 to 2020
When data are exchangeable, the previously discussed conditional
independence condition would generally hold.
Bernoulli Likelihood
For binary data y (e.g., coin
flip, pass/fail, diagnosed/not), an intuitive way to analyze is to use a
Bernoulli model: P(y=1∣θ)=θP(y=0∣θ)=1−θ,
Multiple Observations
When there are more than one y,
say y1,…,yN, that are
conditionally independent, we have P(y1,…,yN∣θ)=N∏i=1P(yi∣θ)=θ∑Ni=1yi(1−θ)∑Ni=1(1−yi)=θz(1−θ)N−z,
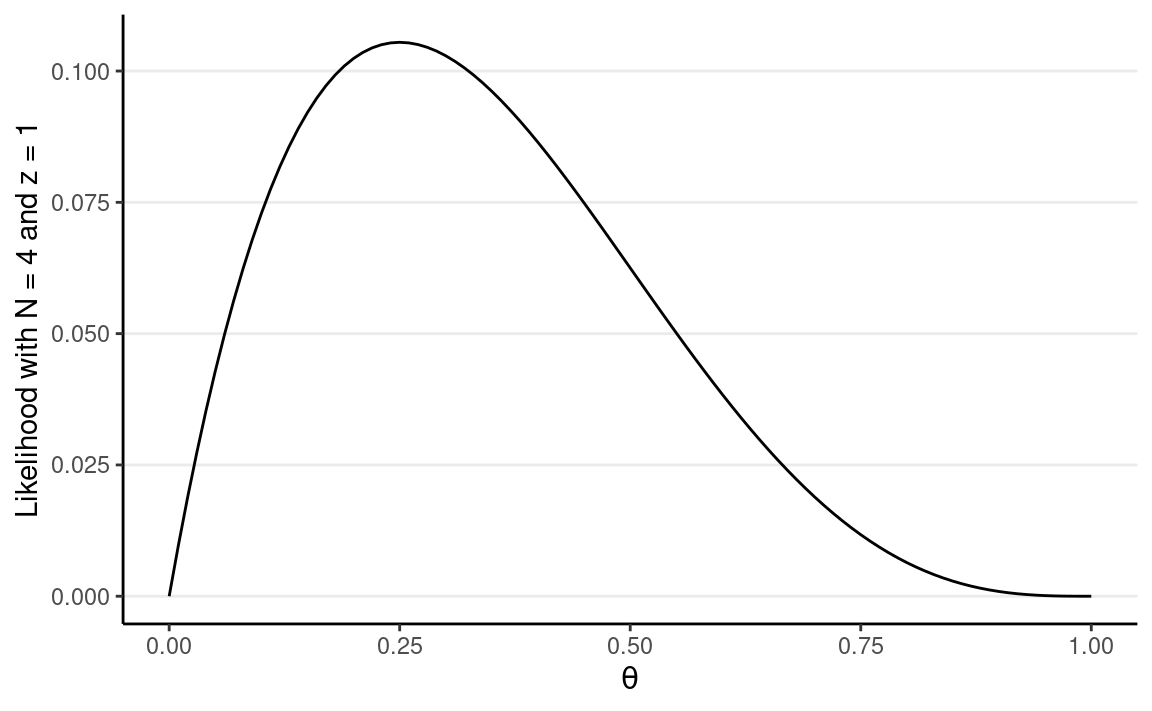
Let’s say N = 4 and z = 1. We can plot the likelihood in R:
# Write the likelihood as a function of theta
lik <- function(th, num_flips = 4, num_heads = 1) {
th ^ num_heads * (1 - th) ^ (num_flips - num_heads)
}
# Likelihood of theta = 0.5
lik(0.5)
#> [1] 0.0625# Plot the likelihood
ggplot(tibble(th = c(0, 1)), aes(x = th)) +
# `stat_function` for plotting a function
stat_function(fun = lik) +
# use `expression()` to get greek letters
labs(x = expression(theta),
y = "Likelihood with N = 4 and z = 1")
Setting Priors
Remember again the relationship between the prior and the posterior:
P(θ|y)∝P(y|θ)P(θ)
One straight forward, brute-force method is to discretize the parameter space into a number of points. For example, by taking θ = 0, 0.05, 0.10, . . . , 0.90, 0.95, 1.00, one can evaluate the posterior at these 21 grid points.
Let’s use a prior that peaks at 0.5 and linearly decreases to both sides. I assume that θ = 0.5 is twice as likely as θ=0.25 or θ=0.75 to reflect my belief that the coin is more likely to be fair.
# Define a grid for the parameter
grid_df <- tibble(th = seq(0, 1, by = 0.05))
# Set the prior mass for each value on the grid
grid_df$pth <- c(0:10, 9:0) # linearly increasing, then decreasing
# Convert pth to a proper distribution such that the value
# sum to one
grid_df$pth <- grid_df$pth / sum(grid_df$pth)
# Plot the prior
ggplot(grid_df, aes(x = th, y = pth)) +
geom_col(aes(x = th, y = pth),
width = 0.01,
) +
labs(y = expression(P(theta)), x = expression(theta))
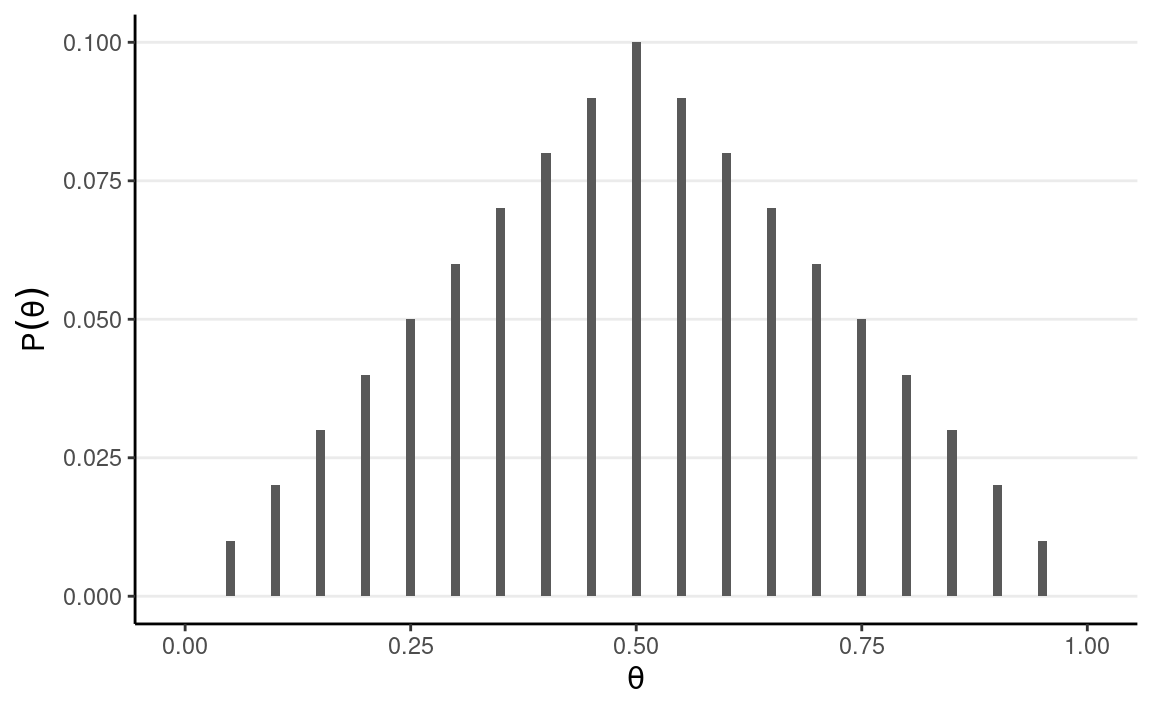
Note the line
grid_df$pth <- grid_df$pth / sum(grid_df$pth), which
ensures that the probability values sum to one. This is a trick we will
use to obtain the posterior probability.
Prior Predictive Distribution
One way to check whether the prior is appropriate is to use the prior predictive distribution. Bayesian models are generative in the sense that they can be used to simulate data. The prior predictive distribution can be obtained by first simulating some θ values from the prior distribution and then simulating a data set for each θ.
# Draw one theta
num_trials <- 4 # number of draws
sim_th1 <- sample(grid_df$th, size = 1,
# based on prior probability
prob = grid_df$pth)
# Simulate new data of four flips based on model
sim_y1 <- rbinom(num_trials, size = 1, prob = sim_th1)
# Repeat many times
# Set number of simulation draws
num_draws <- 1000
sim_th <- sample(grid_df$th, size = num_draws, replace = TRUE,
# based on prior probability
prob = grid_df$pth)
# Use a for loop
# Initialize output
sim_y <- matrix(NA, nrow = num_trials, ncol = num_draws)
for (s in seq_len(num_draws)) {
# Store simulated data in the sth column
sim_y[, s] <- rbinom(num_trials, size = 1, prob = sim_th[s])
}
# Show the first 10 simulated data sets based on prior:
sim_y[, 1:10]
#> [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
#> [1,] 1 0 0 1 0 1 0 0 0 1
#> [2,] 0 0 0 1 1 0 1 0 1 1
#> [3,] 1 0 0 1 0 0 0 1 1 1
#> [4,] 1 0 1 1 1 0 0 1 1 0# Show the distribution of number of heads
sim_heads <- colSums(sim_y)
ggplot(tibble(z = sim_heads), aes(x = z)) +
geom_bar()
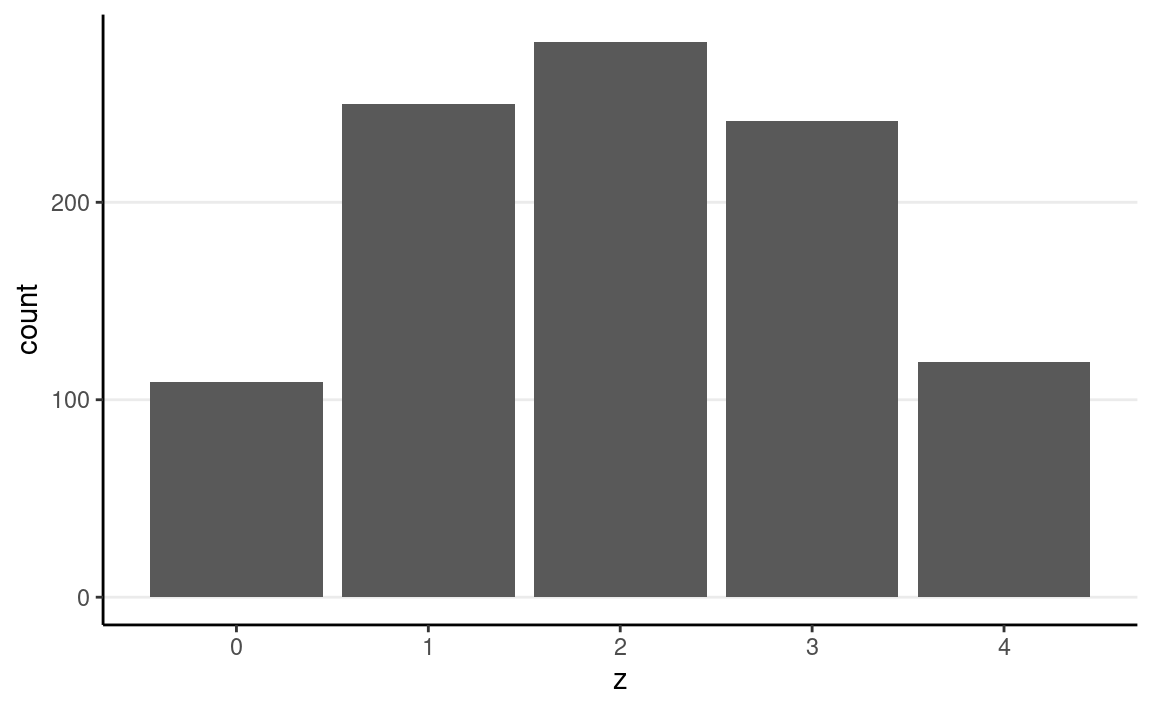
The outcome seems to fit our intuition that it’s more likely to be half heads and half tails, but there is a lot of uncertainty.
Summarizing the Posterior
grid_df <- grid_df %>%
mutate(
# Use our previously defined lik() function
py_th = lik(th, num_flips = 4, num_heads = 1),
# Product of prior and likelihood
`prior x lik` = pth * py_th,
# Scaled the posterior
pth_y = `prior x lik` / sum(`prior x lik`)
)
# Print a table
knitr::kable(grid_df)
| th | pth | py_th | prior x lik | pth_y |
|---|---|---|---|---|
| 0.00 | 0.00 | 0.0000000 | 0.0000000 | 0.0000000 |
| 0.05 | 0.01 | 0.0428687 | 0.0004287 | 0.0073359 |
| 0.10 | 0.02 | 0.0729000 | 0.0014580 | 0.0249500 |
| 0.15 | 0.03 | 0.0921188 | 0.0027636 | 0.0472914 |
| 0.20 | 0.04 | 0.1024000 | 0.0040960 | 0.0700927 |
| 0.25 | 0.05 | 0.1054688 | 0.0052734 | 0.0902416 |
| 0.30 | 0.06 | 0.1029000 | 0.0061740 | 0.1056525 |
| 0.35 | 0.07 | 0.0961187 | 0.0067283 | 0.1151381 |
| 0.40 | 0.08 | 0.0864000 | 0.0069120 | 0.1182815 |
| 0.45 | 0.09 | 0.0748688 | 0.0067382 | 0.1153071 |
| 0.50 | 0.10 | 0.0625000 | 0.0062500 | 0.1069530 |
| 0.55 | 0.09 | 0.0501187 | 0.0045107 | 0.0771891 |
| 0.60 | 0.08 | 0.0384000 | 0.0030720 | 0.0525695 |
| 0.65 | 0.07 | 0.0278687 | 0.0019508 | 0.0333832 |
| 0.70 | 0.06 | 0.0189000 | 0.0011340 | 0.0194056 |
| 0.75 | 0.05 | 0.0117188 | 0.0005859 | 0.0100268 |
| 0.80 | 0.04 | 0.0064000 | 0.0002560 | 0.0043808 |
| 0.85 | 0.03 | 0.0028687 | 0.0000861 | 0.0014727 |
| 0.90 | 0.02 | 0.0009000 | 0.0000180 | 0.0003080 |
| 0.95 | 0.01 | 0.0001187 | 0.0000012 | 0.0000203 |
| 1.00 | 0.00 | 0.0000000 | 0.0000000 | 0.0000000 |
# Plot the prior/likelihood and the posterior
p1 <- ggplot(data = grid_df, aes(x = th)) +
geom_col(aes(x = th - 0.005, y = pth, fill = "prior"),
width = 0.01,
) +
geom_col(aes(x = th + 0.005, y = py_th / sum(py_th),
fill = "scaled likelihood"), width = 0.01,
) +
labs(fill = NULL, y = NULL, x = expression(theta)) +
theme(legend.position = "top")
p2 <- ggplot(data = grid_df, aes(x = th)) +
geom_col(aes(x = th, y = pth_y), width = 0.01) +
labs(
fill = NULL, y = NULL, title = "Posterior",
x = expression(theta)
)
gridExtra::grid.arrange(p1, p2)
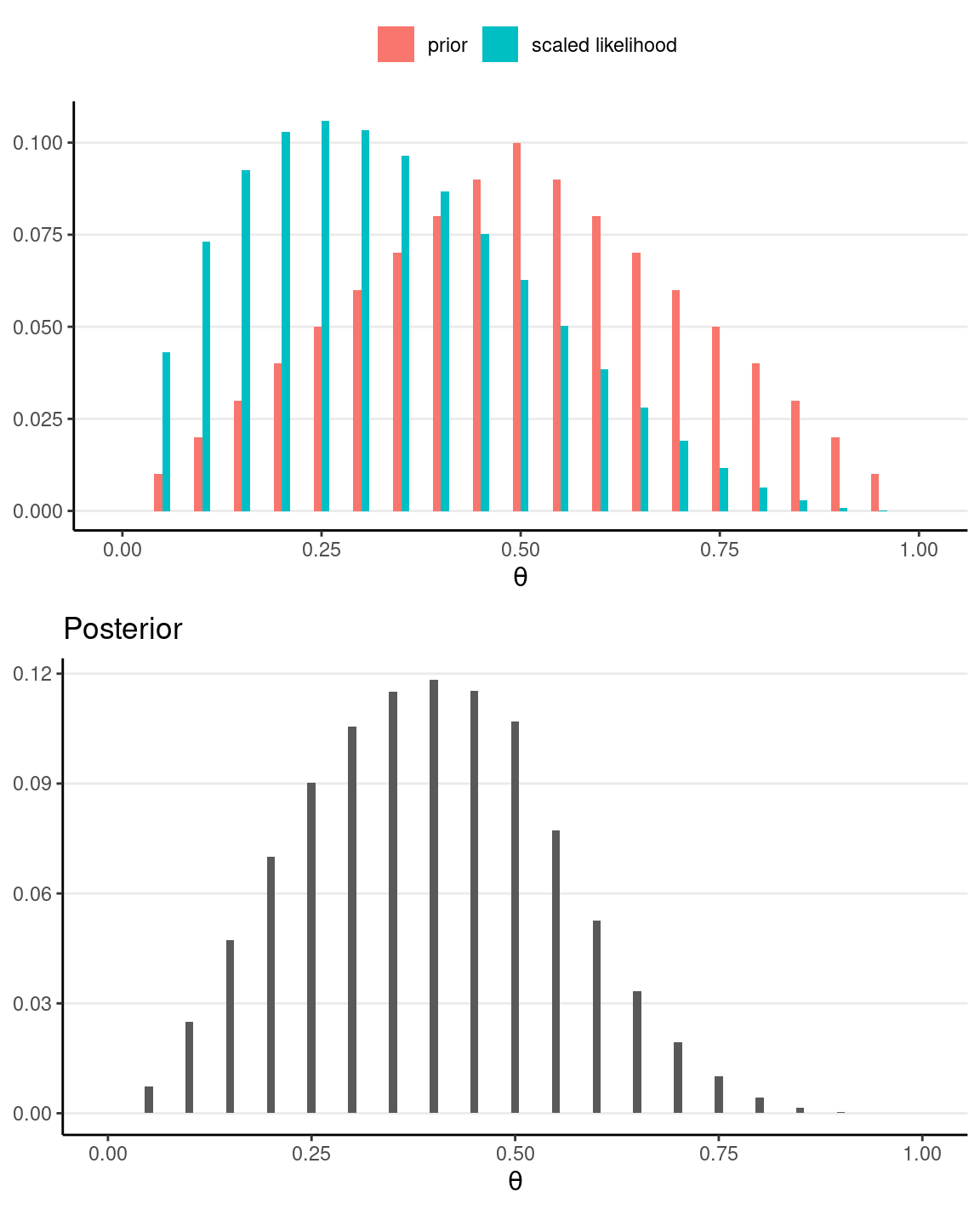
The second graph above shows the posterior distribution, which represents our updated belief about θ. We can summarize it by simulating θ values from it and compute summary statistics:
# Define a function for computing posterior summary
summ_draw <- function(x) {
c(
mean = mean(x),
median = median(x),
sd = sd(x),
mad = mad(x),
`ci.1` = quantile(x, prob = .1, names = FALSE),
`ci.9` = quantile(x, prob = .9, names = FALSE)
)
}
# Sample from the posterior
post_samples <- sample(
grid_df$th,
size = 1000, replace = TRUE,
prob = grid_df$pth_y
)
summ_draw(post_samples)
#> mean median sd mad ci.1 ci.9
#> 0.3848000 0.4000000 0.1538429 0.1482600 0.2000000 0.6000000Influence of Sample Size
If, instead, we have N = 40 and z = 10, the posterior will be more similar to the likelihood.
grid_df2 <- grid_df %>%
mutate(
# Use our previously defined lik() function
py_th = lik(th, num_flips = 40, num_heads = 10),
# Product of prior and likelihood
`prior x lik` = pth * py_th,
# Scaled the posterior
pth_y = `prior x lik` / sum(`prior x lik`)
)
# Plot the prior/likelihood and the posterior
p1 <- ggplot(data = grid_df2, aes(x = th)) +
geom_col(aes(x = th - 0.005, y = pth, fill = "prior"),
width = 0.01,
) +
geom_col(aes(x = th + 0.005, y = py_th / sum(py_th),
fill = "scaled likelihood"), width = 0.01,
) +
labs(fill = NULL, y = NULL, x = expression(theta)) +
theme(legend.position = "top")
p2 <- ggplot(data = grid_df2, aes(x = th)) +
geom_col(aes(x = th, y = pth_y), width = 0.01) +
labs(
fill = NULL, y = NULL, title = "Posterior",
x = expression(theta)
)
gridExtra::grid.arrange(p1, p2)
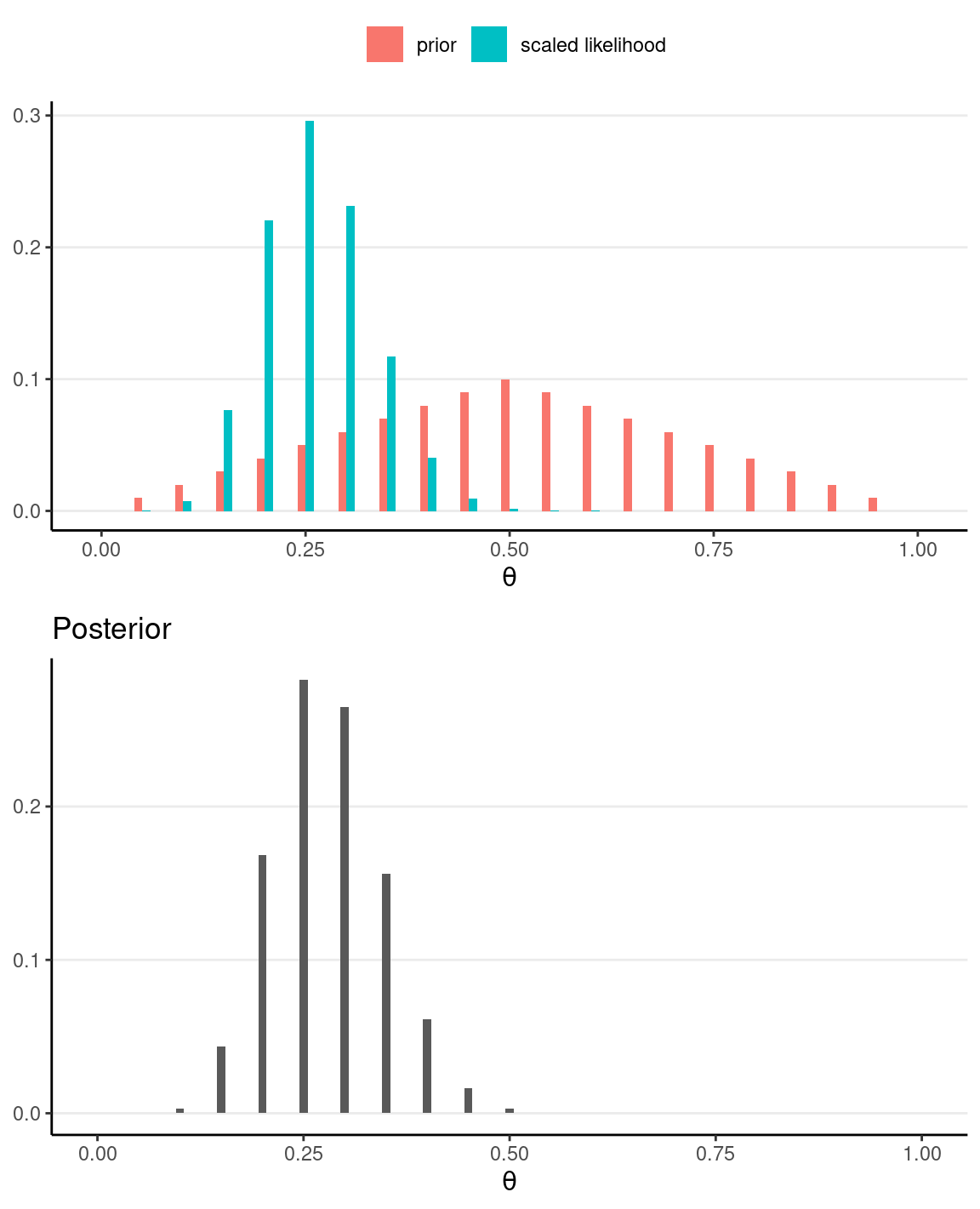
# Sample from the posterior
post_samples <- sample(
grid_df2$th,
size = 1000, replace = TRUE,
prob = grid_df2$pth_y
)
summ_draw(post_samples)
#> mean median sd mad ci.1 ci.9
#> 0.28085000 0.30000000 0.06542215 0.07413000 0.20000000 0.35000000Influence of Prior
If we have a very strong prior concentrated at θ = .5, but still with N = 40 and z = 10, the posterior will be more similar to the prior.
grid_df3 <- grid_df %>%
mutate(
# stronger prior
pth = pth ^ 3,
# scale the prior to sume to 1
pth = pth / sum(pth),
# Use our previously defined lik() function
py_th = lik(th, num_flips = 4, num_heads = 1),
# Product of prior and likelihood
`prior x lik` = pth * py_th,
# Scaled the posterior
pth_y = `prior x lik` / sum(`prior x lik`)
)
# Plot the prior/likelihood and the posterior
p1 <- ggplot(data = grid_df3, aes(x = th)) +
geom_col(aes(x = th - 0.005, y = pth, fill = "prior"),
width = 0.01,
) +
geom_col(aes(x = th + 0.005, y = py_th / sum(py_th),
fill = "scaled likelihood"), width = 0.01,
) +
labs(fill = NULL, y = NULL, x = expression(theta)) +
theme(legend.position = "top")
p2 <- ggplot(data = grid_df3, aes(x = th)) +
geom_col(aes(x = th, y = pth_y), width = 0.01) +
labs(
fill = NULL, y = NULL, title = "Posterior",
x = expression(theta)
)
gridExtra::grid.arrange(p1, p2)
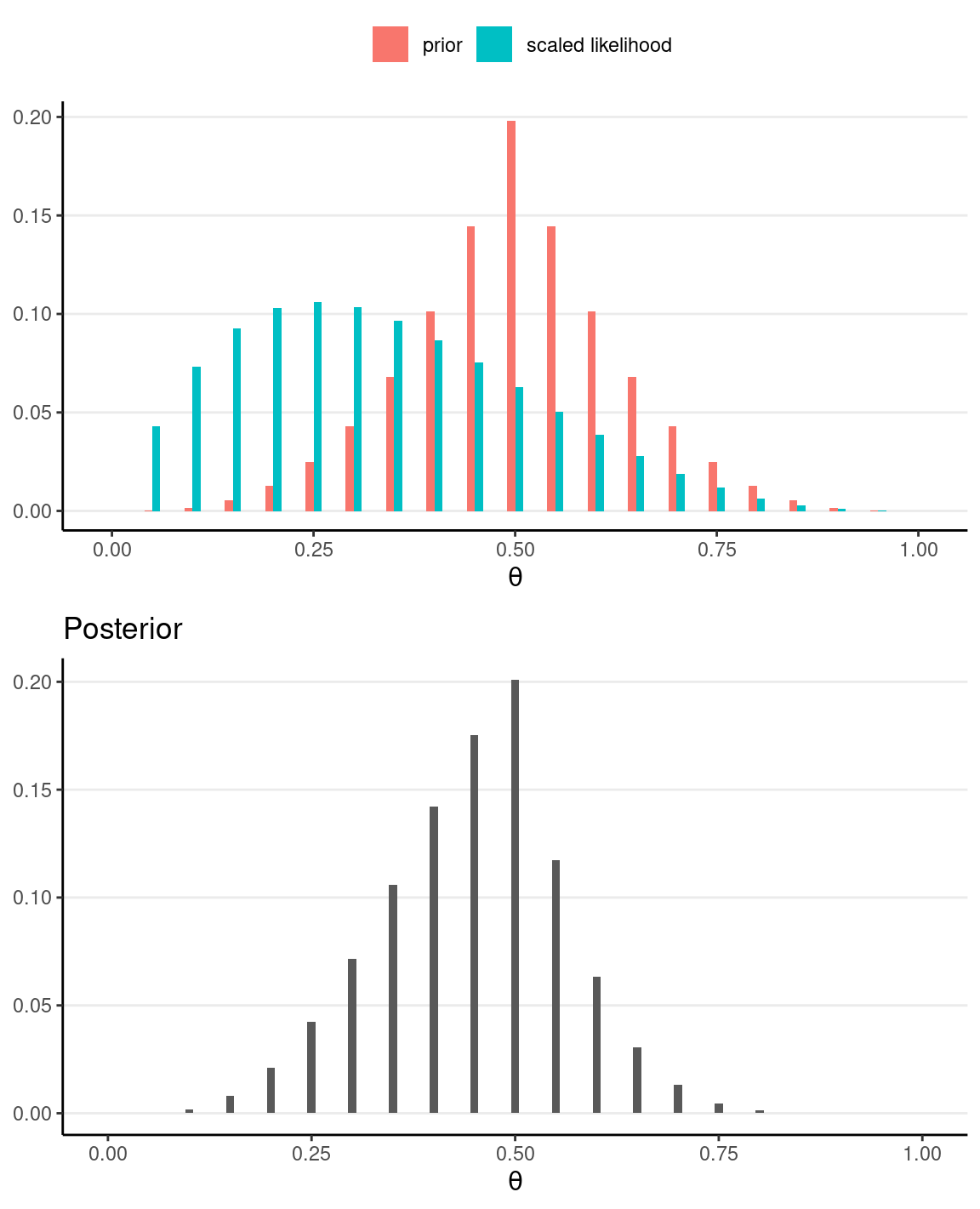
# Sample from the posterior
post_samples <- sample(
grid_df3$th,
size = 1000, replace = TRUE,
prob = grid_df3$pth_y
)
summ_draw(post_samples)
#> mean median sd mad ci.1 ci.9
#> 0.4493000 0.4500000 0.1096656 0.0741300 0.3000000 0.6000000Remark on Grid Approximation
In this note, we discretized θ into a finite number of grid points to compute the posterior, mainly for pedagogical purposes. A big limitation is that our posterior will have no density for values other than the chosen grid points. While increasing the number of grid points (e.g., 1,000) can give more precision, the result is still not truly continuous. A bigger issue is that the computation breaks down when there is more than one parameter; if there are p parameters, with 1,000 grid points per parameter, one needs to evaluate the posterior probability for 1,000p grid points, which is not feasible even with modern computers. So more efficient algorithms, namely Markov chain Monte Carlo (MCMC) methods, will be introduced as we progress in the course.